Content
- Introduction
- Origin
- What is a gene mutation and how do mutations occur?
- How the gene mutation affect heath and Development?
- Lethal & Mutation
- LOF And GOF Mutation
- Software Tools for Genetic Mutation Analysis
- Reference
Meaning: The term was derived from a Greek word 'Muto' which means 'to change. Therefore, mutation may be defined as "any change in the genetic material of organisms which is permanent and heritable from generation to generation
Definition: Process by which sequences of base pairs is altered. Mutation could result in changes in base pairs and in chromosome. The term mutation was given by Hugo de Vries(1901).
What is a gene mutation and how do mutations occur?
- Gene mutation are changes in the sequence of a DNA.
- Gene mutations can be classified in two major ways:
- Hereditary (Germinal):- Germline mutation occur in a parent’s reproductive cell (egg or Sperm). These mutation change the genetic material that the child receives from their parent. You can inherit germline mutation from either parent.
- Aquired (Somatic):- Somatic mutation are a change to a person’s DNA that occurs after conception to any cell that is not germ cell. Somatic mutations don’t pass from parent’s to their child and happen randomly without the mutation existing in a person’s family history. They also can’t pass to future generations.
- To function correctly, each cell depends on thousands of proteins to do their jobs in the right places at the right times.
- Sometimes, gene variants prevent one or more proteins from working properly. By changing a gene's instructions for making a protein, a variant can cause a protein to not to be produced at all.
- When a variant alters a protein that plays a critical role in the body, it can disrupt normal development or cause a health condition.
- A condition caused by variants in one or more genes is called a genetic disorder.
- Gens themselves do not cause disease-Genetic disorders are caused by variants that eliminate a gene's function. Example when people say that someone has "the cystic fibrosis gene" they are usually referring to a version of the CFTR gene that contain a variant that causes the disease. All people including those without cystic fibrosis, have version of the CFTR gene.
- Lethal mutation is a type of mutation in which Lethal means that something has an intense enough impact to cause the death of an organism. So a lethal mutation is a kind of mutation in which the effect(s) might cause mortality or greatly lower the predicted lifespan of an organism bearing the mutation.
- Are all mutations lethal? No, Some mutations may have little to no effect on the organism it impacts. Others mutations may cause disabilities to the organism, whether physically or mentally. This is known as a non-lethal mutation. However, some mutations are quite lethal.
- A classic example of an allele that affects survival is the lethal yellow allele, a spontaneous mutation in mice that makes their coats yellow. This allele was discovered around the turn of the 20th century by the French geneticist Lucien Cuenót, who noticed that it was inherited in an unusual pattern.
- When yellow mice were crossed with normal brown mice, they produced half yellow and half brown offspring. This suggested that the yellow mice were heterozygous, and that the yellow allele, A^Y, was dominant to the brown allele, A. But when two yellow mice were crossed with each other, they produced yellow and brown offspring in a ratio of 2:1, and the yellow offspring did not breed true (were heterozygous). Why was this the case?
- • As it turned out, this unusual ratio reflected that some of the mouse embryos (homozygous (A^y A^y) genotype) died very early in development, long before birth. In other words, at the level of eggs, sperm, and fertilization, the color gene segregated normally, resulting in embryos with a 1:2:1 ratio of (A^y A^y), (A^y A), and (AA) genotypes. However, the (A^y A^y) mice died as tiny embryos, leaving a 2:1 genotype and phenotype ratio among the surviving mice.
- Alleles like A^y, which are lethal when they're homozygous but not when they're heterozygous, are called recessive lethal alleles.
- Sickle Cell anemia
- Cystic Fibrosis
- Achondroplasia
- Familial Hypercholesteramia
- Huntington Disease
- Neurofibromatosis
- Marfan Syndrome
- Cuénot and Baur discovered these first recessive lethal genes because they altered Mendelian inheritance ratios.
- Recessive lethal genes can code for either dominant or recessive traits, but they do not actually cause death unless an organism carries two copies of the lethal allele.
- Examples of human diseases caused by recessive lethal alleles include cystic fibrosis, sickle-cell anemia, and achondroplasia,
- Achondroplasia is an autosomal dominant bone disorder that causes dwarfism. While the inheritance of one achondroplasia allele can cause the disease, the inheritance of two recessive lethal alleles is fatal.
- Dominant lethal genes are expressed in both homozygotes and heterozygotes. But how can alleles like this be passed from one generation to the next if they cause death?
- Dominant lethal genes are rarely detected due to their rapid elimination from populations. One example of a disease caused by a dominant lethal allele is Huntington's disease.
- Huntington's disease
- A neurological disorder in humans, which reduces life expectancy.
- Because the onset of Huntington's disease is slow, individuals carrying the allele can pass it on to their offspring. This allows the allele to be maintained in the population. Dominant traits can also be maintained in the population through recurrent mutations.
- an organism lives normally under one set of conditions, but when certain changes are introduced in its environment, lethality results.
- Favism is a sex-linked, inherited condition that results from deficiency in an enzyme called glucose-6-phosphate dehydrogenase.
- It is most common among people of Mediterranean, African, Southeast Asian, and Sephardic Jewish descent.
- The disease was named because when affected individuals eat fava beans, they develop hemolytic anemia, a condition in which red blood cells break apart and block blood vessels.
- Blockage can cause kidney failure and result in death.
- Semilethal or Sublethal Genes.
- the lethal gene is carried on the sex chromosome, usually X.
- Hemophilia is a hereditary disease caused by deficiencies in clotting factors, which results in impaired blood clotting and coagulation.
- Because the allele responsible for hemophilia is carried on the X chromosome, affected individuals are predominantly males, and they inherit the allele from their mothers.
- The alleles responsible for hemophilia are thus called semilethal or sublethal genes, because they cause the death of only some of the individuals or organisms with the affected genotype.
- Normally, clotting factors help form a temporary scab after a blood vessel is injured to prevent bleeding, but hemophiliacs cannot heal properly after injuries because of their low levels of blood clotting factors.
- Therefore, affected individuals bleed for a longer period of time until clotting occurs. This means that normally minor wounds can be fatal in a person with hemophilia.
- Synthetic lethality describes the genetic interaction between two genes. If either gene is mutated by itself, the organism remains viable. The combination of a mutation in both genes is incompatible with viability and results in lethality.
- Scientists studying the fruit fly observed that pairwise combinations of some mutant alleles were not viable, whereas singly, the same mutant alleles did not cause death (Boone et al., 2007). In other words, some mutations are only lethal when paired with a second mutation. These genes are called synthetic lethal genes.
- When an allele causes lethality, this is evidence that the gene must have a critical function in an organism.
- The discoveries of many lethal alleles have provided information on the functions of genes during development.
- So scientists used conditional and synthetic lethal alleles to study the physiological functions and relationships of genes under specific conditions.
- Gene gain and loss are prevalent in the genomes of diverse organisms and contribute to genetic variation.
- The importance of gene loss (or pseudogene formation) has been almost entirely ignored for a long time, mainly because pseudogenes may not produce full length proteins and are regarded as putatively non-functional.
- the ‘‘less is more’’ hypothesis proposes that gene loss may be an adaptive evolutionary process that is beneficial to organisms.
- Two major mechanisms can cause gene loss: physical removal events (recombination or the mobilization of transposable or viral elements) that lead to the fragment deletion of one or more genes, and deleterious mutations at gene coding regions that cause loss function (LOF) mutations.
- An LOF mutation was first observed in the 5S DNA gene of Xenopus laevis. This gene could not encode a functional 5S rRNA and was named a pseudogene.
- Many pseudogenes caused by LOF mutations were reported in Escherichia coli, yeast, mammals and plants.
- There are four major genetic variations that can lead to LOF mutation,
- First, a nonsense SNP may lead to a premature stop codon, producing a truncated protein sequence. For example, in Arabidopsis, FRIGIDA (FRI) alleles with premature stop codons explain a large fraction of flowering-time variation.
- Second, a SNP that occurs at a canonical splice site may affect splicing. Specifically, a SNP that occurs in a splice donor site causes intron retention in the mRNA, whereas a SNP that occurs in a splice acceptor site removes an exon from the original mRNA. Splice site variations may eventually lead to frameshifts or premature stop codons. In the Arabidopsis relative Capsella rubella, splice site mutations in the FLOWERING LOCUS C gene cause a premature stop codon that promotes flowering.
- Third, insertion or deletion variants with non-integral multiples of 3 located in the gene coding region can lead to frameshifts by disrupting the full length transcript.
- Fourth, the loss of an initiation codon can lead to LOF mutations. The loss of transcription start codon (ATG) variations prevents gene transcription if there is no alternative start codon near the mutation. For example, in humans, the loss of the start codon in the FRMD7 gene leads to idiopathic infantile nystagmus disease.
- LOF mutations have been studied in diverse species at the population level. Almost every genome contains many LOF mutations in either heterozygous or homozygous states. Most LOF mutations in a genome tend to be present at low allele frequencies and have a heterozygous status.
- In humans, an analysis of 7597 genomes identified 17 764 stop-gain variants and 13 915 frameshift variants within 11 369 protein-coding genes.
- Based on 1432 whole exome sequences from five isolated European populations, 173 homozygous LOF mutations were identified within 167 genes.
- Analysis of whole genomes sequencing from 2636 Icelanders and chip-genotype data from 101 584 additional individuals identified 6795 autosomal LOF variants within 4924 gene.
- Our recent study of 1071 A. thaliana genomes from worldwide accessions revealed 60 819 LOF variants within 12 907 genes.
- Compared with functional protein-coding genes (Figure ), LOF mutations are natural gene knockouts that can provide insight into gene function in diverse organisms,
- In rice, a natural LOF mutation in GSE5, which encodes a plasma membrane-associated protein, contributes to grain size diversity.
- In A. thaliana, natural brx LOF alleles confer root adaptation to acidic soil, and a natural knockout allele of ARMADILLO REPEATCONTAINING KINESIN1 causes root hair branching,
- In addition, different LOF mutations of duplicated genes in diverse A. thaliana populations can lead to hybrid incompatibility,
- Essential genes are those genes that are necessary for survival.
- An individual cannot survive if LOF mutations have occurred in essential genes; otherwise, these genes would be considered non-essential.
- LOF mutations in the genomes of natural populations provide direct evidence to delimit essential genes.
- Our essential gene number is much larger but understandable. Because growth conditions in laboratories are much better than in natural habitats, plants probably tolerate more LOF mutations when grown in laboratories. Therefore, the definition of essential genes is context dependent, reflecting the niche that the organisms inhabit.
- LOF mutations in the genome may be neutral, deleterious, or advantageous. Neutral or less-deleterious LOF mutations can be tolerated and may even accumulate during range expansion.
- the ‘‘less is more’’ hypothesis proposes that gene loss may be beneficial to organisms. Adaptive LOF mutations have been observed frequently in bacteria and yeast. An analysis of bacterial fitness in more than 100 different conditions revealed that LOF mutations can provide fitness benefits.
- LOF mutations play important roles in the evolution and diversification of diverse organisms. In plants, a premature stop codon in GL4 caused smaller grain size and loss of seed shattering during African rice domestication.
- Similarly, flower color plays an important role in pollinator attraction (Bradshaw and Schemske, 2003). In Petunia axillaris, an LOF mutation in ANTHOCYANIN2 (AN2) occurred independently at least five times, changing the flower color from violet-red to white compared with Petunia integrifolia and influencing the shift in pollinator attraction from bees to hawkmoths.
- LOF mutations have also been found to be beneficial in human evolution. A CASP12 LOF allele is known to promote resistance to severe sepsis, and rare LOF mutations in SLC30A8 can protect against type 2 diabetes.
- Species-wide studies could be performed to understand the genome-wide distribution patterns, functional effects, and evolutionary importance of LOF mutations. Several studies of LOF mutations at the genome level have been performed in natural populations.
- In particular, our recent study revealed that the level of nucleotide diversity, the density of transposable elements, and gene family size are positively correlated with the presence of LOF mutations.
- the evolutionary effects of natural LOF mutations on other genes in the same pathway may be complicated. When genes become non-functional, other genes in the same pathway may accumulate LOF mutations as well.
- the functional effects of LOF mutations are interesting to study. Genome-wide study of LOF variants can provide information on gene lethality based on the frequency of LOF alleles for a given gene in natural environments.
- In yeast S. cerevisiae, a study of 1106 essential gene knockouts found that 88 (9%) of them could survive through adaptive evolution, and these were defined as evolvable essential genes.
- Synthetic biology can offer interesting insights into the beneficial effects of regulator loss. For example, in Chinese hamster ovary cells, repressor loss in the promoter region of PuroR leads to high gene expression and drug resistance.
- the agricultural importance of LOF mutations is largely unknown. In plants, several studies have reported that LOF mutations can also increase crop yield. For example, LOF of GW2, a gene that encodes a RING-type E3 ubiquitin ligase, can increase grain width and weight.
- Similarly, an LOF mutation of MEI2-LIKE PROTEIN4 (OML4) leads to large and heavy grains in rice (Oryza sativa).
- Gain-of-function mutations-cause either new function or function expressed at new times or location within organism.
- Gain of function mutation is dominant. Single mutation could derive cell toward cancer.
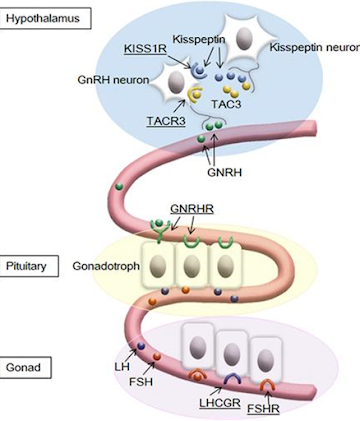
- Known major G-protein–coupled receptors (GPCRs) and their ligands involved in the hypothalamus-pituitarygonadal axis. GPCRs are underlined. A gain-of-function mutation in KISS1R has been identified in a patient with precocious puberty.
- GOF studies are typically applied in virology and have revealed many details regarding the biological mechanisms responsible for viral transmission and replication.
- The high replication and mutation rates of viruses often lead to escape mutants. It is common for these new viral lineages to acquire genomic changes that reduce or eliminate the affinity of natural or vaccine-induced antibodies toward the virus.
- early studies regarding the E484K mutation of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein suggest affinity towards the angiotensinconverting enzyme 2 (ACE2) receptor, the target of the virus, is enhanced. Simultaneously, neutralization by serum antibodies sourced from patients who had previously recovered from wild-type SARS-CoV-2 infection is evaded more effectively.
- The U.S. Food and Drug Administration (FDA) requires animal testing on vaccines before human trials can begin. Since viral tropism towards the model species is unlikely to exist already, in cases where human viruses are under investigation, strains that can infect the model species must be generated. This can be achieved using GOFR, wherein the virus is passage through the animal, thereby allowing molecular determinants of transmissibility to be identified and vaccines under investigation to be tested.
- Technological advancement has indeed contributed its quota in bioinformatics. Analysis of gene’s constituent for mutation can be done using software tools.
- The accuracy of these tools differs from one to the other. Here are some these tools considered in the review: Biomedical Mutation Analysis (BMA), MutaNET, VariantMaster, Mutation Surveyor, Mutation taster, Polyphen2, SIFT, KGGSeq, Galaxy, Ga TK, AlaMut, Imutant, SNPEffect, HOPE etc.
- BMA was designed for an accessible analysis of mutations online. BMA is a user friendly application by which the user compare aligned sequences with a reference sequence.
- The output given will be the place where the changes are and to what it changed. Assists in computing changes in nucleotide and amino acid sequences.
- BMA is an online application that stores all information related to the mutation analyses. This tool performs thanks to an analysis algorithm able to evaluate multiple patients, where each one can include multiple sequences.
- Mutations are the main source of variation in organisms of a species.
- Mutations provide a path for evolution of new varieties and/or species.
- Mutations have helped much in understanding the structure and function of gene. Practical
- Some mutations have beneficial effects and are useful in crop improvement. Mutations in both qualitative and quantitative traits have been exploited in plant breeding Examples:-
- Two amber grain coloured mutants viz., Sharabati sonara and 'Pusa lerma' have been produced in India from 'Sonara 64' and 'Lerma rajo' varieties of Mexican origin.
- Using X-rays irradiation, semi dwarf varieties of rice have been produced. Till 1982 more than 45 useful varieties produced through mutation were in use.
- Many more varieties in pulses, maize, Sorghum spices etc. have been produced through mutation breeding.
- Early and late ripening varieties of wheat and rice have been developed by mutation.
- Some varieties of maize, wheat, soyabean and other crops developed through mutation have been reported to be rich in protein and free amino acids like lysine.
- Through mutations, resistance of various diseases have been raised in various crops.
- Mutation in a gene allow the gene to be studied in greater detail.
- Mutation is helpful in establishing the relationship between a gene and the protein produced by the gene.
- Biochemical mutations help in deciphering the biochemical pathway into individual reaction steps. Such a pathway can never be studied without mutational studies.
- The process is generally random and unpredictable.
- Useful mutants recessive are rare and predominantly recessive.
- Mutants can have strong negative pleiotropic effects on other traits.
- Health risks: handling, chemical mutagens; radiations, fast neutrons treatments.
- Most mutants are of no use to breeding even if a large number of mutants can be produced.
- Loewe, L., Genetic Mutation | Learn Science at Scitable. 2008,https://www.nature.com/scitable/ topicpage/geneticmutation-1127?error=cookies_not_supported&code= f4b6c104-de21-4785-987b-84d03ed7b087
- Brennan, J., How Can a Mutation in DNA Affect Protein Synthesis? 2018, https://sciencing.com/can-mutationdnaaffect-protein-synthesis-2028.ht
- Khan S and Vihinen M., Performance of protein stability predictors. - PubMed - NCBI. 2019 https://www.ncbi.nlm.nih.gov/pubmed/20232415
- Genetics Home Reference, What is a gene mutation and how do mutations occur? 2019, https://ghr.nlm.nih.gov/primer/mutationsanddisorders/gen emutation
- Bartee, L., How Gene Mutations Occur – Mt Hood Community College Biology 102. 2016, https://openoregon.pressbooks.pub/mhccbiology102/chapt,


















No comments:
Post a Comment